Upstage AI Lab 2기
2024년 1월 2일 (화) Day_015 ~ 1월 11일 (목) Day_022
EDA 조별 프로젝트
1. 데이터셋 선정
https://www.kaggle.com/datasets/alexteboul/diabetes-health-indicators-dataset/data
- Diabetes_012 : 0 = no diabetes 1 = prediabetes 2 = diabetes
- HighBP : 0 = no high BP 1 = high BP
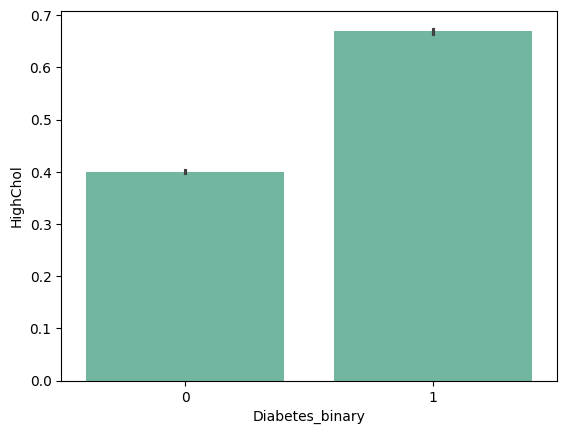
- HighChol : 0 = no high cholesterol 1 = high cholesterol
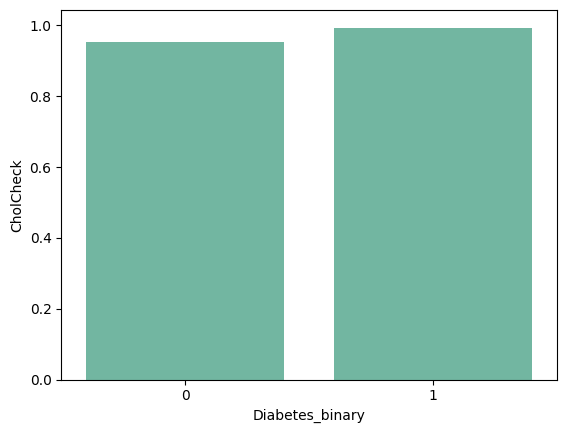
- CholCheck : 0 = no cholesterol check in 5 years 1 = yes cholesterol check in 5 years
- BMI
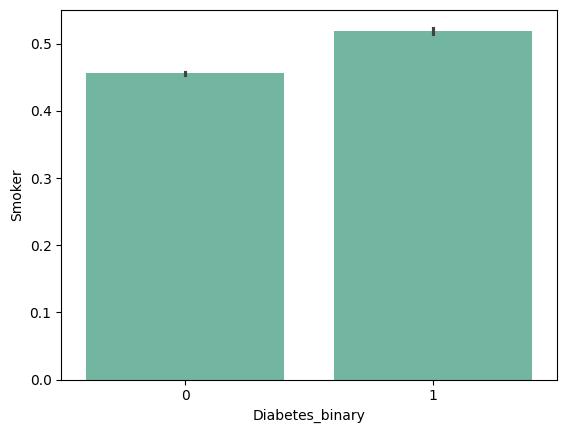
- Smoker : Have you smoked at least 100 cigarettes in your entire life? [Note: 5 packs = 100 cigarettes] 0 = no 1 = yes
- Stroke : (Ever told) you had a stroke. 0 = no 1 = yes
- HeartDiseaseorAttack : coronary heart disease (CHD) or myocardial infarction (MI) 0 = no 1 = yes
- PhysActivity : physical activity in past 30 days - not including job 0 = no 1 = yes
- Fruits : Consume Fruit 1 or more times per day 0 = no 1 = yes
- Veggies : Consume Vegetables 1 or more times per day 0 = no 1 = yes
- HvyAlcoholConsump : Heavy drinkers (adult men having more than 14 drinks per week and adult women having more than 7 drinks per week) 0 = no 1 = yes
- AnyHealthcare : Have any kind of health care coverage, including health insurance, prepaid plans such as HMO, etc. 0 = no 1 = yes
- NoDocbcCost : Was there a time in the past 12 months when you needed to see a doctor but could not because of cost? 0 = no 1 = yes
- GenHlth : Would you say that in general your health is: scale 1-5 1 = excellent 2 = very good 3 = good 4 = fair 5 = poor
- MentHlth : Now thinking about your mental health, which includes stress, depression, and problems with emotions, for how many days during the past 30 days was your mental health not good? scale 1-30 days
- PhysHlth : Now thinking about your physical health, which includes physical illness and injury, for how many days during the past 30 days was your physical health not good? scale 1-30 days
- DiffWalk : Do you have serious difficulty walking or climbing stairs? 0 = no 1 = yes
- Sex : 0 = female 1 = male
- Age : 13-level age category (_AGEG5YR see codebook) 1 = 18-24 9 = 60-64 13 = 80 or older
- Education : Education level (EDUCA see codebook) scale 1-6 1 = Never attended school or only kindergarten 2 = Grades 1 through 8 (Elementary) 3 = Grades 9 through 11 (Some high school) 4 = Grade 12 or GED (High school graduate) 5 = College 1 year to 3 years (Some college or technical school) 6 = College 4 years or more (College graduate)
- Income : Income scale (INCOME2 see codebook) scale 1-8 1 = less than $10,000 5 = less than $35,000 8 = $75,000 or more
3 csv filies in the dataset : 'Diabetes_012', 'Diabetes_binary', 'Diabetes_binary_5050'
'Diabetes_012' / 'Diabetes_binary' : 같은 데이터, 다른 레이블. sample n = 253680 (253680 rows × 22 columns)
('Diabetes_012' : 0 = no diabetes, 1 = prediabetes, 2 = diabetes
/ 'Diabetes_binary ' : 0 = no diabetes, 1 = prediabetes or diabetes )
 |
 |
'Diabetes_binary_5050' : 'no diabetes' vs. ' diabetes' 를 50:50으로 resampled.
2024년 1월 3일 10시 회의 결과
1. binary 불균형 데이터('Diabetes_binary')로 분석
2. 중복행 제거
- 변수 대부분이 범주형 데이터이고, 'BMI', 'MentHlth', 'PhysHlth' 등은 numerical data이긴 하나 discrete data이므로 모든 value가 겹치는 행은 유의하지 않은 데이터일 것으로 판단.
diabetes_binary = pd.read_csv('.\data\diabetes\diabetes_binary_health_indicators_BRFSS2015.csv')
df_diabetes_binary = diabetes_binary.astype(int)
df_diabetes_binary = df_diabetes_binary.drop_duplicates()중복행 제거 결과 : 229474 rows × 22 columns
(no diabetes : 194377 / diabetes : 35097)
columns
['Diabetes_binary', 'HighBP', 'HighChol', 'CholCheck', 'BMI', 'Smoker', 'Stroke', 'HeartDiseaseorAttack', 'PhysActivity', 'Fruits', 'Veggies', 'HvyAlcoholConsump', 'AnyHealthcare', 'NoDocbcCost', 'GenHlth', 'MentHlth', 'PhysHlth', 'DiffWalk', 'Sex', 'Age', 'Education', 'Income']
- dependent variable : 'Diabetes_binary'
- binary data : 'HighBP', 'HighChol', 'CholCheck', 'Smoker', 'Stroke', 'HeartDiseaseorAttack', 'PhysActivity', 'Fruits', 'Veggies', 'HvyAlcoholConsump', 'AnyHealthcare', 'NoDocbcCost', 'DiffWalk', 'Sex'
- categorical data : 'GenHlth', 'Age', 'Education', 'Income'
- numeric / disrete data : 'BMI', 'MentHlth', 'PhysHlth'
'Diabetes_binary'의 category name 컬럼을 추가
diab_mappping = {
0 : 'no diabetes',
1 : 'diabetes'
}
df_diabetes_binary['Diabetes_bin_cat'] = np.select([df_diabetes_binary['Diabetes_binary'] == key for key in diab_mappping], diab_mappping.values())
barplots
| 'HighBP' | 'HighChol' | 'CholCheck' | 'BMI' |
 |
 |
 |
 |
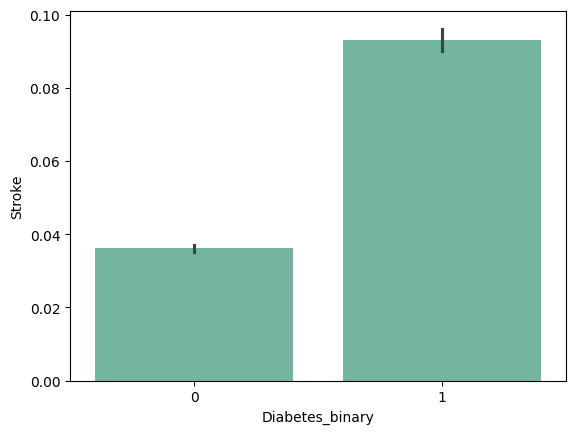
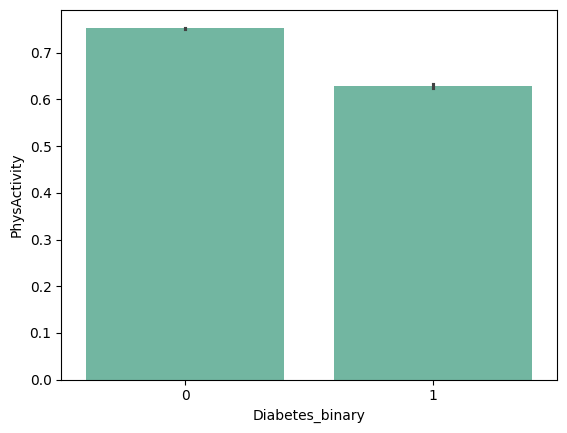
| 'Smoker' | 'Stroke' | 'HeartDiseaseorAttack' | 'PhysActivity' |
 |
 |
 |
 |
| 'Fruits' | 'Veggies' | 'HvyAlcoholConsump' | 'AnyHealthcare' |
 |
 |
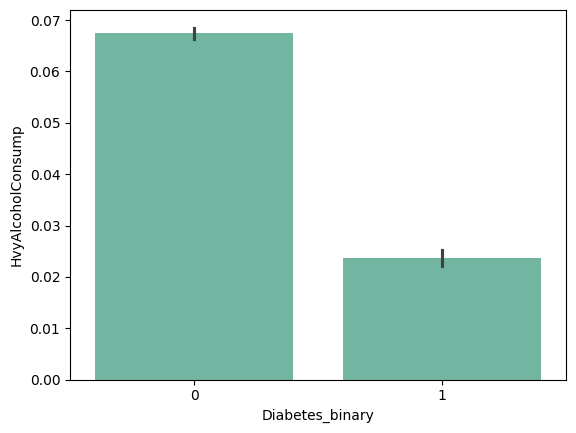 |
 |
| 'NoDocbcCost' | 'GenHlth' | 'MentHlth' | 'PhysHlth' |
 |
 |
 |
 |
| 'DiffWalk' | 'Age' | 'Education' | 'Income' |
 |
 |
 |
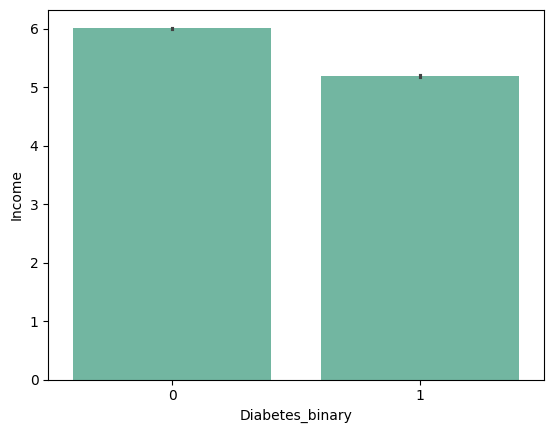 |
from barplot results :
+ ) 'HighBP', 'HighChol', 'Stroke', 'HeartDiseaseorAttack', 'PhysHlth' , 'DiffWalk'
- ) 'HvyAlcoholConsump'???

df_diabetes_binary.corr()
plt.figure(figsize=(20, 16))
sns.heatmap(df_diabetes_binary.corr(), annot=True, fmt='.2f', cmap='Blues')
카이제곱 검증
주어진 변수 중 categorical data인 변수들만 선택하여 카이제곱 검증한 결과
'GenHlth', 'Age', 'Education', 'Income', 'BMI', 'MentHlth', 'PhysHlth' 변수에 대한 p_value 모두 0.05 이하
chi_squred_results = []
# categorical
# dep : 'Diabetes_binary'
# indep : 'GenHlth', 'Age', 'Education', 'Income', 'BMI', 'MentHlth', 'PhysHlth'
for column in df_diabetes_binary[['Age', 'BMI', 'GenHlth', 'MentHlth', 'PhysHlth', 'Education', 'Income']]:
contingency_table = pd.crosstab(df_diabetes_binary[column], df_diabetes_binary['Diabetes_binary'])
chi2, p, _, _ = chi2_contingency(contingency_table)
chi_squred_results.append({
'Variable' : column,
'Chi-squared' : chi2,
'P-value' : f'{p:.8f}'
})
chi2_results_df = pd.DataFrame(chi_squred_results)
chi2_results_df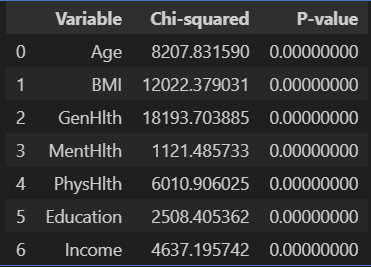
Logistic Regression
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report, accuracy_score# binary
# dep : 'Diabetes_binary'
# indep : 'HighBP', 'HighChol', 'Smoker', 'Stroke', 'HeartDiseaseorAttack', 'PhysActivity', 'Fruits', 'Veggies', 'HvyAlcoholConsump', 'DiffWalk', 'Sex'
indep_vars = ['HighBP', 'HighChol', 'Smoker', 'Stroke', 'HeartDiseaseorAttack', 'PhysActivity', 'Fruits', 'Veggies', 'HvyAlcoholConsump', 'DiffWalk', 'Sex']
X = df_diabetes_binary[indep_vars]
y = df_diabetes_binary['Diabetes_binary']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
logreg_model = LogisticRegression()
f1_results = []
for column in X.columns:
logreg_model.fit(X_train[[column]], y_train)
y_pred_temp = logreg_model.predict(X_test[[column]])
f1_temp = f1_score(y_test, y_pred_temp)
f1_results.append({
'Variable' : column,
'F1 Score' : f'{f1_temp :.4f}'
})
f1_results_df = pd.DataFrame(f1_results)
f1_results_df
| True Positives | False Positives |
| False Negatives | True Negatives |
precision = (True Positive) / ( True Positives + False Positives )
recall = (True Positive) / ( True Positives + False Negatives )
F1 score = precision 와 recall 의 조화평균
(1 = perfect precision and recall
0 = either precision or recall are zero)
before feedback. accuracy score를 썼음
# binary
# dep : 'Diabetes_binary'
# indep : 'HighBP', 'HighChol', 'Smoker', 'Stroke', 'HeartDiseaseorAttack', 'PhysActivity', 'Fruits', 'Veggies', 'HvyAlcoholConsump', 'DiffWalk', 'Sex'
indep_vars = ['HighBP', 'HighChol', 'Smoker', 'Stroke', 'HeartDiseaseorAttack', 'PhysActivity', 'Fruits', 'Veggies', 'HvyAlcoholConsump', 'DiffWalk', 'Sex']
X = df_diabetes_binary[indep_vars]
y = df_diabetes_binary['Diabetes_binary']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
logreg_model = LogisticRegression()
accuracy_results = []
for column in X.columns:
logreg_model.fit(X_train[[column]], y_train)
y_pred_temp = logreg_model.predict(X_test[[column]])
accuracy_temp = accuracy_score(y_test, y_pred_temp)
accuracy_results.append({
'Variable' : column,
'Accuracy' : f'{accuracy_temp:.4f}'
})
accuracy_results_df = pd.DataFrame(accuracy_results)
accuracy_results_df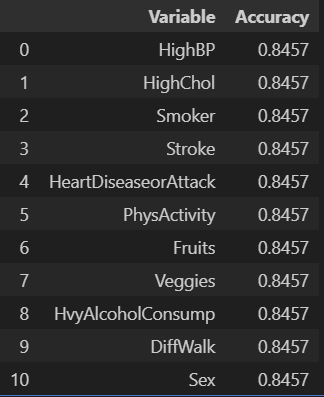
note. class가 균등하지 않기 때문에 accuracy score가 아닌 f1-score를 써야 함.
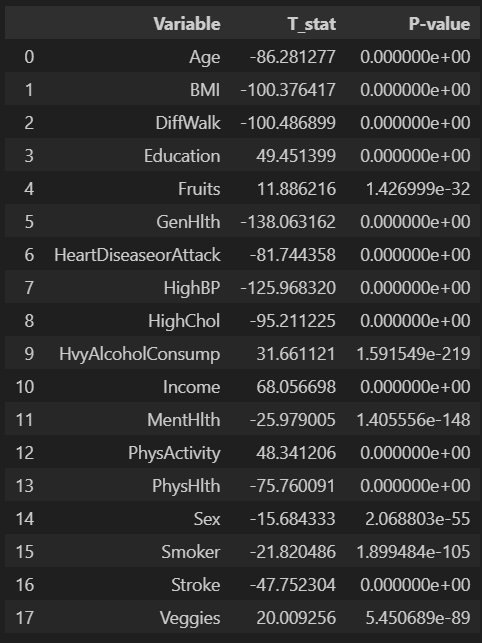
wrong. t-test를 쓸 수 있는 데이터가 아님.
pt_all = pd.pivot_table(data=df_diabetes_binary, index='Diabetes_binary', values=df_diabetes_binary.columns[1:], aggfunc='mean')
pt_all.drop(columns=['AnyHealthcare', 'CholCheck', 'NoDocbcCost'])
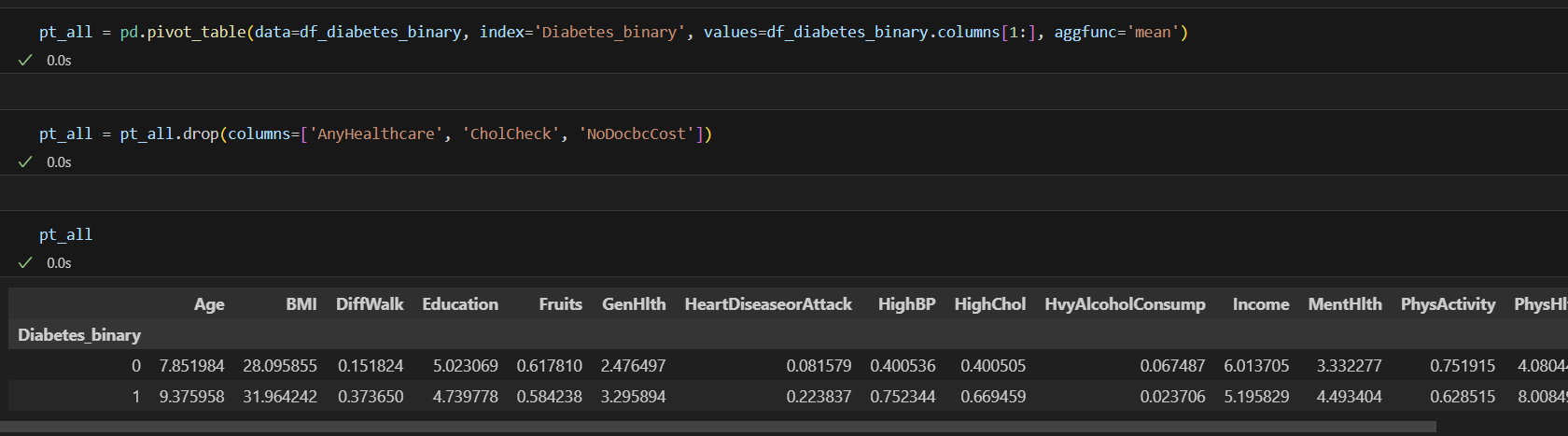
ttest_results = []
for column in pt_all.columns:
group0 = df_diabetes_binary[df_diabetes_binary['Diabetes_binary'] == 0][column]
group1 = df_diabetes_binary[df_diabetes_binary['Diabetes_binary'] == 1][column]
t_stat, p_value = ttest_ind(group0, group1)
ttest_results.append({
'Variable' : column,
'T_stat' : t_stat,
'P-value' : f'{p_value:.8f}'
})
ttest_results_df = pd.DataFrame(ttest_results)
ttest_results_df
2:40pm 강사님 피드백
research questions가 명확한 데이터셋
당뇨병을 예측하는 것이 목표인 데이터셋이라 머신러닝까지 가야할 수도 있음.
(예를 들어 XGBoost로 feature importance 추출, 혹은 linear model 학습)
- Can survey questions from the BRFSS provide accurate predictions of whether an individual has diabetes?
- What risk factors are most predictive of diabetes risk?
- Can we use a subset of the risk factors to accurately predict whether an individual has diabetes?
- Can we create a short form of questions from the BRFSS using feature selection to accurately predict if someone might have diabetes or is at high risk of diabetes?
현재 데이터셋은 class imbalance가 심하지 않아 별도의 처리과정은 필요하지 않을 것 같음.
어떤 목표로 진행하고 싶은가에 따라 추후 진행해야할 분석이 달라질 것임.
예측성능을 높이고 싶다면 XGBoost나 LightGBM을 써볼 수 있지만, 변수별 설명력은 떨어질 수 있음.
train / test set을 별도의 csv 파일로 나눠 저장해서 조원들간 공유하는 것이 좋은 방법일 수 있음.
EDA의 이상적 목표
스토리라인이 중요. 어떤 business implication이 있을지, research question들을 세부화하여 주제를 정하고 진행하는 것.
+ 종속 변인과 독립 변인의 연관성이 크다는 것을 보여줄 수 있는 분포의 차이가 있는지
+ CDC에서 같이 결합해서 볼 수 있는 데이터가 있는지 찾아보는 것도 추천
주형님 피드백
통계분석으로 1차 feature selection, LightGBM으로 feature importance를 추출
(다른 데이터 분석할 때 10-fold validation을 해봤었는데 그 때는 tree마다 feature importance가 다르게 나오는 문제가 있었음. -> feedback : tuning을 잘 하면 변동성을 줄일 수 있음. feature importance 가 직관적이고 영향력을 잘 보여줄 수는 있음.)
추가 작업 : 'Diabetes_binary_5050'
sample n = 69057
0 = no diabetes, 1 = prediabetes or diabetes.
(no diabetes : 33960, diabetes : 35097)
상관계수 정렬
corr_matrix = df_diabetes_binary_5050.corr()
upper_tri = corr_matrix.where(np.triu(np.ones(corr_matrix.shape), k=1).astype(np.bool_))
corr_series = upper_tri.unstack()
sorted_corr_series = corr_series.sort_values(ascending=False)
Diabetes_binary 와 양의 상관관계가 높은 변수들
GenHlth : 0.396571
HighBP : 0.372048
BMI : 0.285643
HighChol : 0.281399
Age : 0.274550
DiffWalk : 0.267082
Diabetes_binary 와 음의 상관관계가 높은 변수들
PhysActivity : -0.150281
Education : -0.158522
Income : -0.212846
상호간 상관관계가 높아서 통합가능성이 있는 변수들
PhysHlth GenHlth 0.550138
DiffWalk PhysHlth 0.484092
DiffWalk GenHlth 0.472338
Income Education 0.450376
PhysHlth MentHlth 0.376625
Age HighBP 0.333721
MentHlth GenHlth 0.310093
HighChol HighBP 0.308987
GenHlth HighBP 0.308459
GenHlth HeartDiseaseorAttack 0.271502
GenHlth BMI 0.256642
DiffWalk MentHlth 0.246948
DiffWalk BMI 0.240667
Age HighChol 0.235779
- 어디까지 cut?

VIF 계산
def calculat_vif(df) :
vif_data = pd.DataFrame()
vif_data['Variable'] = df.columns
vif_data['VIF'] = [variance_inflation_factor(df.values, i) for i in range(df.shape[1])]
return vif_data
vif_result = calculat_vif(df_diabetes_binary)
vif_result
reference :
to be continued....
'Upstage AI Lab 2기' 카테고리의 다른 글
| Upstage AI Lab 2기 [Day015-022] EDA 조별 프로젝트 (데이터 개요) (1) | 2024.01.04 |
|---|---|
| Upstage AI Lab 2기 [Day015-022] EDA 조별 프로젝트 (1) 기초통계 (0) | 2024.01.04 |
| Upstage AI Lab 2기 [Day014] (2) EDA 실습 (0) | 2023.12.31 |
| Upstage AI Lab 2기 [Day014] (1) Seaborn_기초 (0) | 2023.12.29 |
| Upstage AI Lab 2기 [Day012] (2) pandas_기초_1 (0) | 2023.12.28 |



